Replications for introduction to g-methods: marginal structural models (part 2)
Published:
This tutorial series aims to replicate g-methods explained in this paper by Naimi, A. I., Cole, S. R., and Kennedy, E. H (2017)1 using R. Originally, the paper used SAS to demonstrate g-methods.
In this tutorial, we will focus on replicating the results using marginal structural models to estimate the average causal effects of always taking treatment and compared to never-taking treatment. From our knowledge of the data-generating process, we know this average causal effect to be \(50\). In our tutorial, we will pay more attention to computation rather than proofs to perform replciations.
Reminder: Our Settings
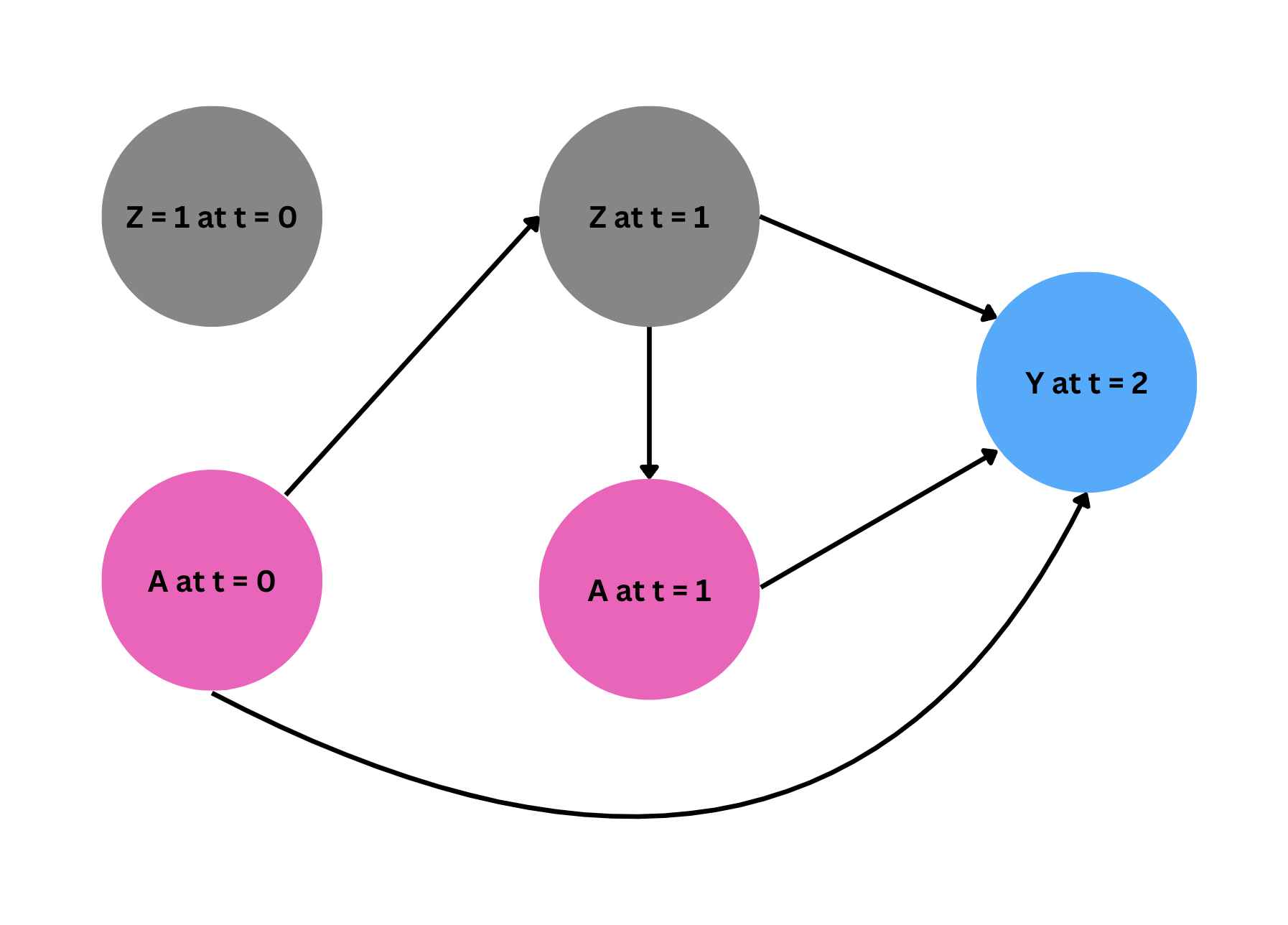 Under the identifying assumptions described in the paper, we will estimate the average causal effect of always taking treatment ($a_0 = 1, a_1 = 1$), compared to never taking treatment ($a_0 = 0, a_1 = 0$) in both time periods. For notation, we are using subscripts to indicate time periods.
Under the identifying assumptions described in the paper, we will estimate the average causal effect of always taking treatment ($a_0 = 1, a_1 = 1$), compared to never taking treatment ($a_0 = 0, a_1 = 0$) in both time periods. For notation, we are using subscripts to indicate time periods.Marginal Structural Models
Marginal structural models typically relate a marginal summary (e.g., average) of potential outcomes to the treatment and parameter of interest ($\beta$). Inverse probability weighting is most commonly used to estimate the parameters of these models. Specifically, to estimate $\beta$, we calculate the predicted probabilities of the observed treatments at each time $t$, given prior covariates and treatments. By weighting the observed data by the inverse of these probabilities, we generate a ``pseudo-population’’ in which treatment at each time $t$ is no longer related to, and thus no longer confounded by, prior covariates and treatments. Consequently, weighting a parametric model for the outcome by the inverse probability of treatment allows us to account for the fact that both the time-varying covariates and the treatment affect subsequent treatment statuses.
Naimi, A. I., Cole, S. R., & Kennedy, E. H. (2017). An introduction to g methods. International journal of epidemiology, 46(2), 756-762. ↩
